New model would be used to analyze biopsies of salivary glands Using artificial intelligence (AI) to analyze biopsies of the salivary glands in people with Sjögren’s disease may help identify patients who are likely to have disease involvement in vital organs, a new study reports. According to the researchers, the novel model developed by the
Google DeepMind’s AlphaGenome AI predicts how non-coding DNA can drive disease
 Approximately 1% of the human genome encodes proteins. The remaining DNA is non-coding, but still influences the activity of proteins by regulating thousands of genes. These non-coding sequences may play major roles in diseases from cancer to heart disease, and might hold the key to developing novel therapeutic approaches, some with curative potential, for specific subsets of these diseases.
Approximately 1% of the human genome encodes proteins. The remaining DNA is non-coding, but still influences the activity of proteins by regulating thousands of genes. These non-coding sequences may play major roles in diseases from cancer to heart disease, and might hold the key to developing novel therapeutic approaches, some with curative potential, for specific subsets of these diseases.
Google DeepMind’s latest model
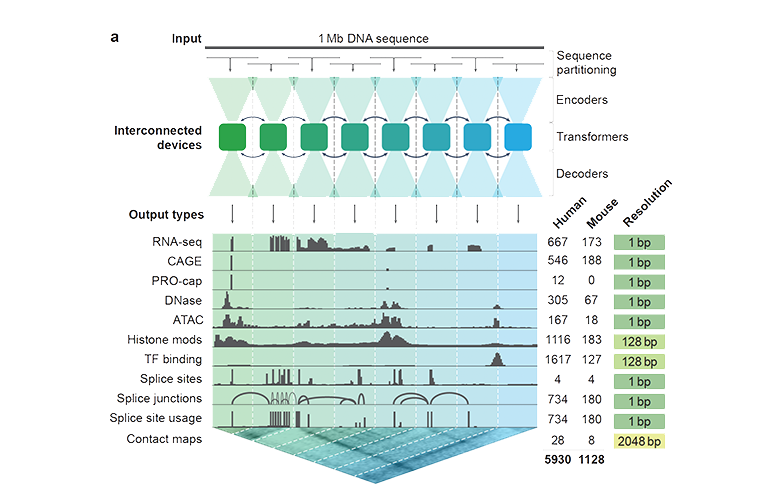
To help explore such potential, Google DeepMind in London has created an AI model, the large-scale, hybrid deep learning model AlphaGenome that could help make sense of the non-coding portions of DNA. The model predicts the expression levels of genes and how they may be affected by mutations that occur over long stretches of DNA. Up to one million DNA letters can be input into the model to receive thousands of predictions. This overcomes a fundamental limitation of previous models, which typically had to trade off between capturing long-range effects and achieving single-letter resolution. The predictions are sensitive to single-letter changes, allowing scientists to predict how mutations will affect gene expression.
The ‘sequence to function’ model takes stretches of DNA and predicts various properties, including the expression level of genes and how they might be affected by mutations.
The model was able to accurately predict non-coding mutations that indirectly activated a nearby gene, which is a driver of a type of leukemia, as identified in previous studies.
The AlphaGenome model was trained on data from humans and mice and has not been tested on other organisms. The model is not perfect; it struggles to identify sequences that alter the expression of a gene more than 100,000 base pairs away. It also does not capture how a cell’s changing nature may affect how DNA sequences function.
It’s a milestone for the field. For the first time, we have a single model that unifies long-range context, base-level precision and state-of-the-art performance across a whole spectrum of genomic tasks —Dr. Caleb Lareau, Memorial Sloan Kettering Cancer Center in an announcement
The video was originally featured in the Google DeepMind post “AlphaGenome: AI for better understanding the genome,” published on June 25, 2025.
Outpacing other models
Previous models, such as Splice AI or ChromBPNet, specialized in a single type of genomic signal. These models have focused on an individual task, such as predicting levels of gene expression or determining how exons code for distinct proteins. In contrast, AlphaGenome predicts across 11 modalities, offering a holistic view of genomic regulation.
AlphaGenome can handle one million DNA letters at single-base-pair resolution, a huge leap in scale and resolution. It is also the first model to jointly predict splice site positions, usage, junctions, and RNA coverage. AlphaGenome outperforms generalist and specialist models on most tasks.
Researchers doing non-commercial work can now access AlphaGenome through DeepMind’s servers. A full release is planned.