Introduction Deep learning is an advanced machine learning technique based on artificial neural networks inspired by a human brain, especially featuring the capability to automatically extract complex patterns of data by multi-layer neural networks [1,2]. The neural network consists of input, hidden, and output layers. Unlike conventional machine learning algorithms, deep learning architectures have multiple
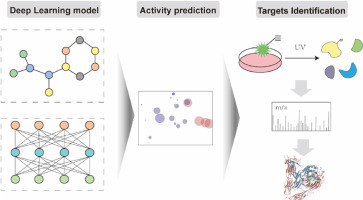
A dual-view deep learning-driven discovery of cinnamoyl anthranilic acid derivatives against …
Introduction
The orthopoxvirus genus comprises double-stranded DNA viruses, including variola virus, vaccinia virus, monkeypox virus, and cowpox virus[1], [2]. Among these, variola virus, the causative agent of smallpox, is particularly notorious for its high contagion and mortality rates. It was declared to be eradicated by the World Health Organization (WHO) in 1980. The monkeypox virus (MPXV), closely related to variola virus, is another transmissible pathogen responsible for mpox. Following outbreaks in 2022 and 2024, mpox has affected over 100,000 individuals globally and has been designated a Public Health Emergency of International Concern twice by the WHO. Three drugs initially authorized for smallpox (Tecovirimat, Cidofovir, and Brincidofovir) are considered potential treatments for mpox; however, concerns regarding drug resistance, renal toxicity, and bioavailability remain unresolved. Therefore, developing effective therapeutic strategies to combat the ongoing orthopoxvirus epidemic is an urgent public health priority[3].
Deep learning has emerged as a powerful tool for automatically mining and analyzing raw data, leading to significant advancements in biomedical research[4], [5]. These models facilitate the exploration of large-scale chemical structures and complex molecular interaction patterns with high flexibility and notable accuracy, thereby accelerating the screening of numerous candidate compounds and development of new therapeutic drugs. A growing body of evidence underscores the potential of artificial intelligence (AI)-aided approaches for anti-infective drug discovery[6]. Consequently, this study aimed to leverage deep learning techniques as a pivotal tool for identifying structurally new candidates with potential efficacy against orthopoxviruses.
Section snippets
Materials
All materials for synthesis were obtained from Bidepharm (China) or Innochem (China). Rabbit anti-ITGB3 monoclonal antibody and anti-rabbit IgG secondary antibody was purchased from Abcam (USA). Streptavidin magnetic beads were obtained from MCE (USA). ITGB3 antibody for functional inhibition assay was obtained from Santa Cruze (USA). Recombinant ITGB3 protein was obtained from HUABIO (Hangzhou China).
Deep learning data collections
The compounds with the anti-orthopoxvirus bioactivity value (IC50 or EC50) of less than 30
Construction of a dual-view deep learning model
In this study, a dual-view deep learning model for discovering novel anti-orthopoxvirus agents was constructed (Figure 1a). First, according to the criteria of established anti-orthopoxvirus bioactivity, the compounds from existing assays of the ChEMBL database[9] were filtered to obtain 1803 compounds with reported anti-orthopoxvirus bioactivity to facilitate the deep learning prediction model of new anti-orthopoxvirus agents.
As for the deep learning framework, we converted these compounds as
Discussion
Integrins represent a class of heterodimeric transmembrane receptors composed of α and β subunits[21]. The β3 subunit, namely ITGB3, is recognized as a member of the β subunit family. It typically binds to the αV subunit to form the αVβ3 complex[24], [25]. Upon binding to integrins on the cell surface, viruses initiate a cascade of downstream signaling pathways, facilitating their entry and invasion into host cells. Compound 6 could fit well in the cavity located between α and β subunits, and
Conclusions
To sum up, a reliable dual-view deep learning model has been established for novel anti-orthopoxvirus agent discovery, and a cinnamoyl anthranilic acid derivative (6) was successfully predicted, with an ideal anti-orthopoxvirus potency both in vitro and in vivo. ITGB3 was confirmed to be one of the direct target proteins of 6 through ABPP technique, and it may be served as a potential target for orthopoxvirus eradication, which deserves further investigations.
CRediT authorship contribution statement
Fan Liu: Methodology, Investigation, Data curation. Mengyi Xu: Writing – original draft, Validation, Data curation. Danqing Song: Supervision, Project administration. Xiaosa Zhao: Software, Methodology, Investigation. Yinghong Li: Validation, Funding acquisition. Yanan Ni: Methodology, Investigation. Yanxiang Wang: Writing – review & editing, Supervision, Project administration, Conceptualization. Weijin Huang: Supervision, Resources. Minghao Yin: Validation, Supervision, Conceptualization.
Conflicts of Interest
The authors declare no competing financial interests.
Ethics statement
This article does not contain any studies with human subjects. The animal study was approved by the Animal Care and Use Committee at the National Institute for Food and Drug Control (NIFDC, Number: 2024 (B) 009).
Declaration of Competing Interest
The authors have declared no conflict of interest
ACKNOWLEDGEMENTS
This research project was supported by grants from Institute of Health and Medicine, Hefei Comprehensive National Science Center Foundation (2024KYQD011), National Natural Science Foundation of China (82373742) and CAMS Initiative for Innovative Medicine (2021-1-I2M-030).
© 2025 Published by Elsevier Masson SAS.