Credit: AI-generated image New research from Carnegie Mellon University's School of Computer Science shows that the smarter the artificial intelligence system, the more selfish it will act. Researchers in the Human-Computer Interaction Institute (HCII) found that large language models (LLMs) that can reason possess selfish tendencies, do not cooperate well with others and can be

Leveraging Machine Learning and Real-World Data to Predict Chronic Obs | COPD
Introduction
Chronic obstructive pulmonary disease (COPD) is a common respiratory condition estimated to affect 4.6–6.6% of American adults1 and is a leading cause of death worldwide.2 The economic and social burden of COPD is considerable, with annual total costs in the USA estimated to be $49 billion and rising.3 Patients with COPD often have substantial comorbidities, placing additional burden on healthcare services and costs of care.3 Treatments that improve the management of COPD and reduce the use of healthcare services can ultimately reduce healthcare expenditures and improve patient quality of life.
Exacerbations of COPD represent a significant cause of morbidity and mortality. Accurately predicting these exacerbations can identify at-risk patients and enable precision therapy, thereby improving health outcomes. Despite research efforts to develop predictive models of COPD exacerbations, they have had limited success. A recent study showed that spirometry predicted future COPD exacerbations;4 however, it is not universally performed in clinical practice and is often performed poorly or may not be accessible to electronic medical review.5 Furthermore, many composite scores, such as ADO (age, dyspnea, obstruction),6 BODE (body mass index, obstruction, dyspnea, and exercise capacity),7 and DOSE (dyspnea, obstruction, smoking status, and exacerbations),8 include spirometry as a key component, which limits their application outside of specialist clinics. In addition, BODE incorporates exercise testing. Some of these composite indices are optimal for mortality and long-term prognosis rather than short-term exacerbation prediction, and some of the indices are more accurate for patients with more severe exacerbations and impairment.
Our current proposed model is designed to be used in a broad patient population, does not require spirometry, and is designed to assess short-to-medium-term exacerbation risk rather than long-term outcomes such as mortality. Composite scores such as ADO,6 BODE,7 and DOSE8 were inconsistent in predicting future exacerbations.9 One risk index, composed of age, percentage of predicted forced expiratory volume in 1 second, oral steroids at entry, cardiovascular comorbidity, and unscheduled clinic/emergency department visits for COPD in the prior year,10 was developed to forecast hospitalization due to COPD exacerbations. However, the study cohort was from a clinical trial for tiotropium and was confined to a single healthcare system, which is unlikely to be generalizable. The cohort was also not externally validated in other independent populations; therefore, this study’s clinical applicability is uncertain. Additionally, a meta-analysis evaluated the ability of current models to predict exacerbations in patients with COPD, concluding that most prediction models were at high risk of bias, and none of the models could be implemented in clinical applications.11
Machine learning (ML) is being more frequently used to predict long-term disease progression in patients with COPD.12 Bayesian Additive Regression Trees (BART) is an ML approach, which compared with other ML algorithms, is more robust to the choice of tuning parameters, yields high prediction accuracy, and provides a full posterior distribution for the predictions allowing for uncertainty quantifications.13
In pilot studies using clinical attributes identified through the COPDGene study, we applied artificial intelligence (AI) and natural language processing (NLP) to both structured and unstructured electronic health record (EHR) data to identify patients at risk of COPD exacerbations.14,15 Building on this foundation and leveraging real-world data (RWD) and BART, we developed a predictive model to identify patients at risk of COPD exacerbations within 24 months of their initial COPD diagnosis. Our model represents an advancement in predictive analytics to enhance patient care and management of COPD.
Study Design and Methods
NLP and COPD Case Confirmation
To independently confirm that our patient case definitions were valid, two independent evaluators (board-certified pulmonologists) examined a representative sample of the cohort (n = 50) to confirm that the patients met our COPD definition and these patients had experienced a previous COPD exacerbation. Additionally, the evaluators also confirmed that a representative sample of the patients who were rejected for not meeting our case definition of COPD was correctly characterized (n = 25).
The study duration was from January 2018 to January 2024. Cases were identified from the Robert Wood Johnson Barnabas Health (RWJBH) System EHRs, using combinations of the following criteria: 1) International Classification of Diseases (ICD) 10th Revision COPD diagnosis code; 2) smoking history or white blood cell/eosinophil/neutrophil count; 3) modified Medical Research Council Dyspnea Scale grade >2; and 4) COPD Assessment Test (CAT) score, pulmonary function test (PFT), or the presence of select comorbidities. The RWJBH system is the largest and most comprehensive healthcare system in New Jersey, comprising 14 healthcare centers (including 12 acute care centers)16,17 and EHRs of approximately 7.5 million individuals. Using an iterative process, initial data pulls were reviewed and the algorithm was refined to remove non-COPD cases. After the algorithm was finalized, a random sample of patients (n = 50) was selected for further review by two clinicians independent of the algorithm development team to confirm the accuracy of the curated data and to ensure the data included specific patient attributes including age, sex, exacerbation history, eosinophil counts and dyspnea severity. Concordance was confirmed in 98% of cases. Deep 6 AI, Inc., provided precision-matching software to mine our EHR data using “structured” and “unstructured” physician notes to identify eligible patients.
Using NLP/AI applied to the structured and unstructured EHR data, we also identified patients with COPD at risk of exacerbation based on clinical features and determinants identified in the COPDGene study.15 Our analysis developed phrases to identify the population at risk of exacerbation. A set of words or “computable phenotypes” was generated based on expert opinion ascertained from NLP/AI query of EHR data using a defined set of elements and logical expressions. The goal was to devise a high-throughput search tool that applies ML/AI to structured EHR fields and unstructured EHR narratives, to rapidly identify and characterize patients with COPD at risk of exacerbation over time.
The Deep 6 AI Cohort Builder™ software was then applied to the Epic EHR of the RWJBH System to initially identify patients with COPD and to subsequently identify patients at risk of exacerbation. The date of the first observed ICD 9th/10th Revision diagnosis for COPD in the EHR was designated as the index date. COPD was identified based on the following list of diagnoses: chronic obstructive airway disease, bronchiolitis obliterans, end-stage chronic obstructive airways disease, chronic obstructive lung disease co-occurrent with acute bronchitis, moderate COPD, severe COPD, bronchitis, chronic pulmonary emphysema, COPD, severe early-onset COPD with acute lower respiratory infection, acute exacerbation of chronic obstructive airways disease, and mild COPD. In parallel, the evaluators also confirmed that some patients included in the cohort had experienced an exacerbation. Moderate and severe exacerbation events were flagged and defined by our case definitions. A moderate exacerbation was defined as an outpatient or emergency room visit with a primary diagnosis for COPD and at least one prescription for a systemic corticosteroid or guideline-recommended antibiotic within 5 days following, or before, the visit; or a physician diagnosis in the EHR identifying a nonhospitalized exacerbation. A severe exacerbation was defined as an inpatient hospitalization with a primary diagnosis of COPD or a physician diagnosis in the EHR identifying a hospitalized exacerbation.
An iterative process identified the population of patients at risk of a COPD exacerbation. This step evaluated the feasibility and extent to which determinants of COPDGene are readily available in the EHR (completeness and accuracy of COPDGene predictor variables). Our evaluators reviewed charts and verified and identified additional data fields not initially captured through NLP. Further verification that patients were at risk of exacerbation involved a random selection of charts from those identified for inclusion. These charts were reviewed by an evaluator (assessing agreement and accuracy). A minimum of 25 cases/charts were also rated by the identified patient matter expert reviewers. To assess inter-rater agreement, Cohen’s kappa statistic and its associated 95% confidence interval were estimated based on the rated cases. Per guidance from Landis and Koch,18 we interpreted our kappa statistic as showing “poor” (<0.00), “slight” (0.00–0.20), “fair” (0.21–0.40), “moderate” (0.41–0.60), “substantial” (0.61–0.80), or “almost perfect” (0.81–1.00) agreement among the raters. All data were examined in a de-identified manner, and the study was deemed to be nonhuman patients research in accordance with the Institutional Review Board of Rutgers University.
Statistical Analyses
All study variables, including baseline and outcome measures, were analyzed descriptively. The predictor variables (sex, gastroesophageal reflux disease [GERD], coronary artery disease, congestive heart failure, cor pulmonale, asthma, dyspnea [none, mild, moderate, severe], smoking [never, past, current], and eosinophil count [≤300 cells/µL, >300 cells/µL]) were reported as a count and a percentage of patients having each predictor. Bivariate associations between each predictor and the occurrence of a COPD exacerbation were computed as odds ratios and 95% confidence intervals.
We then developed multivariable models to predict the occurrence of a COPD exacerbation within 24 months of initial COPD diagnosis. Independent variables included all the variables described above, as well as pack years, comorbidity count, and PFT. To avoid the parametric assumptions of a multivariable logistic regression model (additivity, linearity on log-odds scale), we implemented BART.19,20 BART uses sums of trees to predict the outcome (like gradient boosting) and allows for higher-order interaction terms and nonlinear relationships without specification, compared with other models. BART is a Bayesian approach, and the prior distribution for BART penalizes large trees to prevent overfitting. BART was implemented using the BART package in R. Model performance was determined using C-statistics (area under the receiver operating characteristic [ROC] curve) and ROC curves. Fitting BART deploys a Markov chain Monte Carlo algorithm. At each iteration in the algorithm, and for each tree, decisions are made about which variable(s) to split on. We recorded how often each variable was split as a rough measure of variable importance. We also quantified parameters such as sensitivity, specificity, positive predictive value, and negative predictive value of the algorithm. This was done for predictors of moderate and severe exacerbations separately.
Results
After meeting our pre-determined criteria, 3007 patients were included in the analytic data set. We observed a greater than 95% concordance between evaluators regarding the case definitions of cohort patients with and without COPD and in those patients with COPD who experienced an exacerbation. As shown in Table 1, most patients were approximately 70 years old and male, and 94% were current or former smokers. The most common comorbidities were coronary artery disease (54.6%) and GERD (51.0%). Within 24 months of the index date, approximately 886 patients (29.5%) reported a unique COPD exacerbation. Few patients (<5%) had multiple exacerbations, and those patients were excluded from our analysis (data not shown). Concerning serum biomarkers, a serum eosinophil count was measured in nearly all patients, with approximately 80% of patients having a measurement of ≤300 cells/µL. In patients with multiple eosinophil count measurements, the highest eosinophil count was used for analyses.
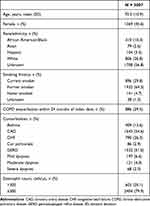 |
Table 1 Summary Statistics for Patient Demographics and Clinical Characteristics |
To determine the bivariate associations between each predictor separately and the risk of COPD exacerbation, we considered an odds ratio of 1.5 to indicate a strong association (vertical line; Figure 1). Strong associations existed between COPD exacerbation and cor pulmonale, moderate dyspnea, severe dyspnea, and number of comorbidities (≥4 vs 0). As expected, smoking history and three or four comorbidities were associated with COPD exacerbations. GERD and coronary artery disease alone were unassociated with COPD exacerbations. Few patients (<2%) failed to have a reported serum eosinophil count, and COPD exacerbations were unassociated with eosinophil counts. Using an unbiased approach, the odds ratio of having a COPD exacerbation was strongly associated with smoking history, dyspnea severity, congestive heart failure, and three or more comorbidities.
 |
Figure 1 Odds ratio estimates of associations between included variables and occurrence of COPD exacerbations. An odds ratio of 1.5 indicates a strong association between the individual predictor and the risk of a COPD exacerbation. Odds ratios of 1.5 or higher are in bold. *All effects were adjusted for age. Abbreviations: CAD, coronary artery disease; CHF, congestive heart failure; CI, confidence interval; COPD, chronic obstructive pulmonary disease; GERD, gastroesophageal reflux disease. |
As a flexible prediction model/ML approach, BART has gained widespread popularity. BART is flexible because it can handle nonlinear main effects and multiway interactions without input from researchers. As shown in Supplementary Figure 1, the frequency by which the BART algorithm is split on each variable is shown in the classification trees as a percentage. The top three most frequently split on variables were eosinophil count, pack years, and moderate dyspnea. Based on the BART model, the ROC is shown in Figure 2. The AUC for our model was 0.69.
 |
Figure 2 ROC curve for the prediction model of COPD exacerbations from the BART model. Abbreviations: AUC, area under the ROC curve; BART, Bayesian Additive Regression Trees; COPD, chronic obstructive pulmonary disease; ROC, receiver operating characteristic. |
Discussion
Using RWD, this study identified that eosinophil count and dyspnea were predictors of COPD exacerbations. This model will enable clinicians to tailor patients’ therapy to improve health outcomes in COPD.
Patient comorbidities in this study are similar to those reported in COPDGene and other longitudinal COPD registries,6,15 although a previous US study using RWD reported a slightly lower mean age and slightly higher proportion of female patients than those reported in our study.21
Accurately predicting COPD exacerbations can identify at-risk patients and can redirect medical management to improve health outcomes.2–4 The most significant risk factor for predicting future exacerbations is a history of prior episodes. However, this knowledge fails as a warning for a first exacerbation, which can often be a pivotal event in a patient’s disease trajectory. Severe exacerbations are particularly concerning, as they are linked to a rapid decline in lung function, with a previous study indicating that approximately 50% of patients may die within 2 years following hospitalization for such an event.22 Although extensive research has been conducted to develop risk scores for predicting COPD exacerbations, such studies are often affected by poor discrimination, inadequate calibration, and a lack of external validation.11 Current predictive models of COPD exacerbations also have shortcomings. Many of the composite measures of disease severity were constructed to characterize quality of life or functional status. These models or tools were not designed to predict acute exacerbations. For example, CAT is a self-administered questionnaire that measures health-related quality of life; the ability of CAT to predict exacerbations is limited. Composite scores like ADO,6 BODE,7 and DOSE8 have also yielded inconsistent findings in predicting future exacerbations.9 Many of these measures were only tested in a few cohorts and were not externally validated in independent populations, so the clinical applicability of these studies is uncertain.
As defined by regulatory agencies, RWD in the medical and healthcare field relate to patient health status or the delivery of healthcare, routinely collected from various sources.23 The growing availability of RWD and the rapid advancement of AI and ML techniques, along with the rising costs and known limitations of traditional trials, have fueled significant interest in leveraging RWD. RWD can enhance the efficiency of clinical research, drive discoveries, and close the evidence gap between clinical research and practice. Concerning predictive models of COPD exacerbation, most tools and composite scores longitudinally require a patient to answer questionnaires or a healthcare provider to administer the test. Such approaches are time-consuming, costly, and instill a bias. Our predictive model using AI/ML can search the EHR unbiasedly to rank predictors of COPD exacerbations without patient or healthcare provider interaction. Using this approach and contemporary statistical analyses, we found that eosinophil count predicted a COPD exacerbation in the next 24 months. This is aligned with a previous systematic review and meta-analysis, which identified that high blood eosinophil levels (>300 cells/μL) could predict the risk of moderate-severe COPD exacerbations in subgroups of patients.24 However, a retrospective study reported that patients with eosinophil levels of ≥300 cells/μL did not show a significant association with the rate of recurrent severe exacerbations.25 We also found that three or more comorbidities (structured data) and moderate dyspnea severity predicted a COPD exacerbation. A previous study also found that comorbidity test index score and a combination of at least one pulmonary disease can predict the risk of moderate-severe acute exacerbations in patients with COPD.26 To the best of our knowledge, there are limited studies which have investigated whether moderate dyspnea can predict a COPD exacerbation.
Unlike other tools, our model using BART predicted a first exacerbation that may identify patients at risk of progressive disease, morbidity, or mortality. BART is an ML method using a nonparametric tree-based approach to predict outcomes from a series of predictor variables. It is similar to classification tree analysis, except rather than using a single large tree, it uses a sum of many small trees. This can be thought of as an ensemble of “weak learners”. BART has advantages over logistic regression in that the model can discover/capture nonlinear relationships and higher-order interactions without input from the analyst. Furthermore, the sum-of-trees models are more stable than single-tree models and lead to smoother functions of the data.27 BART has also performed well in data analysis competitions.28 Our model searches the EHR for parameters that already exist without the need to order tests or perform PFTs. We found that over 95% of patients in our EHR have had a complete blood count that contains an eosinophil count that could be used as a biomarker for predicting an exacerbation (data not shown). This is important since most patients with COPD in the USA are managed by primary care providers who have limited access to sophisticated testing.
Concerning this study, limitations exist regarding our patient cohort and analyses. Patients excluded from this study for lacking 24 months of continuous eligibility in the EHR may differ in their disease profile from those with ≥24 months of eligibility. The study population identified may not be generalizable to other populations in the USA. Our study was conducted retrospectively; a prospective analysis is necessary to validate the predictive algorithm.
Conclusion
Using AI/ML, NLP, and RWD, our study shows that eosinophil count and dyspnea are important predictors of exacerbations in patients with COPD. Our model was able to make these predictions in an unbiased manner and in a broad clinical population encompassing primary care and specialist clinics, and these findings are applicable over a wide clinical severity range. Additionally, we examined a random sample of patients after finalization of the algorithm to verify the accuracy of the curated data and confirm the data included specific patient attributes, with concordance observed in 98% of cases. This helped to ensure robustness and validation of our study findings. Furthermore, these data do not require spirometry or exercise testing, neither of which are widely available outside of specialist clinics. The prognostic value of capturing moderate or severe dyspnea in unstructured data is noteworthy because, in prior studies, dyspnea is usually measured using validated questionnaires not widely used in routine practice. The prognostic value of blood eosinophil counts in a COPD population is consistent with prior studies of COPD exacerbation risk stratification. Although the value of our model will require further prospective validation, these results suggest that active monitoring of eosinophil counts and selected patient-reported experiences of dyspnea in clinical practice may identify patients at risk of exacerbations. This will help guide precision therapy to improve healthcare outcomes of patients with COPD in real-world practice.
Notation of Prior Abstract Publication/Presentation
A summary of these results was presented at the American Thoracic Society 2024 congress: R. A. Panettieri, J. Roy, N. Gontarczyk Uczkowski, A. Tyler, J. Attanucci, T. O’Riordan, K. Wrobleski. Leveraging Machine Learning and Real-world Data to Predict Chronic Obstructive Pulmonary Disease Exacerbation. Poster 724. Available from: https://www.atsjournals.org/doi/abs/10.1164/ajrccm-conference.2024.209.1_MeetingAbstracts.A2801.
Ethics Approval
This study was approved by the Institutional Review Board (IRB) of Rutgers University. This study complied with all applicable laws regarding subject privacy, as described in the Declaration of Helsinki. This study used existing, fully de-identified data from an electronic database that complied with the requirements of the Health Insurance Portability and Accountability Act, and the patient(s) cannot be identified, directly or through identifiers. Additionally, the study results are in tabular form and aggregate analyses that omit patient identification, as per Title 45 of CFR, Part 46. Therefore, the IRB waived the need for informed consent in this study.
Acknowledgments
Editorial support (in the form of writing assistance, including preparation of the draft manuscript under the direction and guidance of the authors, collating and incorporating authors’ comments, grammatical editing, and referencing) was provided by Sarah Case, MSc of Luna, OPEN Health Communications, and funded by GSK, in accordance with Good Publication Practice (GPP) guidelines (www.ismpp.org/gpp-2022).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was funded by GSK (study ID: 217413) following the Good Publication Practice (GPP) guidelines (www.ismpp.org/gpp-2022).
Disclosure
R.A.P. Jr reports research grants from AstraZeneca, Genentech, GSK, and Novartis, and consultant/speaker fees from AstraZeneca, Genentech, Merck, Regeneron, and Sanofi. A.T is an employee of Deep 6 AI, Inc. J.A was an employee of Deep 6 AI, Inc at the time of study. T.G.O’R. and K.K-W. are employees of and hold financial equities in GSK. The authors report no other conflicts of interest in this work.
References
1. Tkacz J, Evans KA, Touchette DR, et al. PRIMUS – prompt initiation of maintenance therapy in the US: a real-world analysis of clinical and economic outcomes among patients initiating triple therapy following a COPD exacerbation. Int J Chron Obstruct Pulmon Dis. 2022;17:329–342. doi:10.2147/copd.S347735
2. Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: 2025 report; 2024. Available from: https://goldcopd.org/2025-gold-report/.
3. Larsen DL, Gandhi H, Pollack M, Feigler N, Patel S, Wise RA. The quality of care and economic burden of COPD in the United States: considerations for managing patients and improving outcomes. Am Health Drug Benefits. 2022;15(2):57–64.
4. Marott JL, Ingebrigtsen TS, Çolak Y, Vestbo J, Nordestgaard BG, Lange P. Predicting exacerbations in COPD in the Danish general population. Respir Med. 2024;224:107557. doi:10.1016/j.rmed.2024.107557
5. Johns DP, Walters JA, Walters EH. Diagnosis and early detection of COPD using spirometry. J Thorac Dis. 2014;6(11):1557–1569. doi:10.3978/j.issn.2072-1439.2014.08.18
6. Puhan MA, Garcia-Aymerich J, Frey M, et al. Expansion of the prognostic assessment of patients with chronic obstructive pulmonary disease: the updated BODE index and the ADO index. Lancet. 2009;374(9691):704–711. doi:10.1016/s0140-6736(09)61301-5
7. Celli BR, Cote CG, Marin JM, et al. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(10):1005–1012. doi:10.1056/NEJMoa021322
8. Jones RC, Donaldson GC, Chavannes NH, et al. Derivation and validation of a composite index of severity in chronic obstructive pulmonary disease: the DOSE Index. Am J Respir Crit Care Med. 2009;180(12):1189–1195. doi:10.1164/rccm.200902-0271OC
9. Bertens LC, Reitsma JB, Moons KG, et al. Development and validation of a model to predict the risk of exacerbations in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2013;8:493–499. doi:10.2147/copd.S49609
10. Niewoehner DE, Lokhnygina Y, Rice K, et al. Risk indexes for exacerbations and hospitalizations due to COPD. Chest. 2007;131(1):20–28. doi:10.1378/chest.06-1316
11. Guerra B, Gaveikaite V, Bianchi C, Puhan MA. Prediction models for exacerbations in patients with COPD. Eur Respir Rev. 2017;26(143):160061. doi:10.1183/16000617.0061-2016
12. Smith LA, Oakden-Rayner L, Bird A, et al. Machine learning and deep learning predictive models for long-term prognosis in patients with chronic obstructive pulmonary disease: a systematic review and meta-analysis. Lancet Digit Health. 2023;5(12):e872–e881. doi:10.1016/s2589-7500(23)00177-2
13. Um S, Linero AR, Sinha D, Bandyopadhyay D. Bayesian Additive Regression Trees for multivariate skewed responses. Stat Med. 2023;42(3):246–263. doi:10.1002/sim.9613
14. Zakusylo A, Wrobelski K, Tyler A, Roy J, O’Riordan T, Panettieri R. Leveraging artificial intelligence to create a chronic obstructive pulmonary disease exacerbation risk algorithm. Chest. 2023;164(suppl 4):A4959–A4960. doi:10.1016/j.chest.2023.07.3213
15. Maselli DJ, Bhatt SP, Anzueto A, et al. Clinical epidemiology of COPD: insights from 10 years of the COPDGene study. Chest. 2019;156(2):228–238. doi:10.1016/j.chest.2019.04.135
16. RWJBarnabas Health. Why RWJBarnabas Health? 2025. Available from: https://www.rwjbh.org/why-rwjbarnabas-health-/.
17. RWJBarnabas Health. Corporate System Fact Sheet; 2024. Available from: https://www.rwjbh.org/documents/RWJBH-CORP-SYS-Fact-Sheet-2024.pdf.
18. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. doi:10.2307/2529310
19. Tan YV, Roy J. Bayesian Additive Regression Trees and the General BART model. Stat Med. 2019;38(25):5048–5069. doi:10.1002/sim.8347
20. Chipman HA, George EI, McCulloch RE. BART: Bayesian Additive Regression Trees. Ann Appl Stat. 2010;4(1):266–298. doi:10.1214/09-AOAS285
21. Young C, Lee LY, DiRocco KK, et al. Adherence and persistence with single-inhaler triple therapy among patients with COPD using commercial and Medicare Advantage US health plan claims data. Adv Ther. 2025;42(2):830–848. doi:10.1007/s12325-024-03055-w
22. Connors AF Jr, Dawson NV, Thomas C, et al. Outcomes following acute exacerbation of severe chronic obstructive lung disease. The SUPPORT investigators (Study to Understand Prognoses and Preferences for Outcomes and Risks of Treatments). Am J Respir Crit Care Med. 1996;154(4 Pt 1):959–967. doi:10.1164/ajrccm.154.4.8887592
23. FDA. Real-world evidence; 2024. Available from: https://www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence.
24. Chen F, Yang M, Wang H, Liu L, Shen Y, Chen L. High blood eosinophils predict the risk of COPD exacerbation: a systematic review and meta-analysis. PLoS One. 2024;19(10):e0302318. doi:10.1371/journal.pone.0302318
25. Adir Y, Hakrush O, Shteinberg M, Schneer S, Agusti A. Circulating eosinophil levels do not predict severe exacerbations in COPD: a retrospective study. ERJ Open Res. 2018;4(3):00022–2018. doi:10.1183/23120541.00022-2018
26. Chen Q, Wang X, Yao X, Zhang L, Liu X. COTE and pulmonary comorbidities predict moderate-to-severe acute exacerbation and hospitalization in COPD. Int J Chron Obstruct Pulmon Dis. 2025;20:1893–1913. doi:10.2147/copd.S518218
27. Hill J, Linero A, Murray J. Bayesian Additive Regression Trees: a review and look forward. Annu Rev Stat Appl. 2020;7(1):251–278. doi:10.1146/annurev-statistics-031219-041110
28. Dorie V, Hill J, Shalit U. Automated versus do-it-yourself methods for causal inference: lessons learned from a data analysis competition. Statist Sci. 2019;34(1):43–68. doi:10.1214/18-STS667



