Introduction Deep learning is an advanced machine learning technique based on artificial neural networks inspired by a human brain, especially featuring the capability to automatically extract complex patterns of data by multi-layer neural networks [1,2]. The neural network consists of input, hidden, and output layers. Unlike conventional machine learning algorithms, deep learning architectures have multiple
Tractography from T1-weighted MRI: Empirically exploring the clinical viability of streamline …
Cai, Leon Y., Lee, Ho Hin, Johnson, Graham W., Newlin, Nancy R., Ramadass, Karthik, Kim, Michael E., Archer, Derek B., Hohman, Timothy J., Jefferson, Angela L., Begnoche, J. Patrick, Boyd, Brian D., Taylor, Warren D., Morgan, Victoria L., Englot, Dario J., Nath, Vishwesh, Chotai, Silky, Barquero, Laura, D’Archangel, Micah, Cutting, Laurie E., Dawant, Benoit M., Rheault, François, Moyer, Daniel C., & Schilling, Kurt G. (2024). Tractography from T1-weighted MRI: Empirically exploring the clinical viability of streamline propagation without diffusion MRI. *Imaging Neuroscience, 2*, 1-20. https://doi.org/10.1162/imag_a_00259
Over the last few decades, diffusion MRI (dMRI) streamline tractography has become the main way to estimate white matter (WM) pathways—the brain’s wiring—while a person is alive. But a big limitation is that this method usually needs a special type of scan called high angular resolution diffusion imaging (HARDI), which can be hard to get during regular medical care. This means tractography is mostly used in research settings and with certain groups of patients, limiting its use in everyday clinical practice and for rare or underfunded diseases. Because of this, having a tractography method that works with common clinical scans would be very important. Such a method would need to perform flexible tractography, use only standard clinical imaging as input, and be openly available for anyone to use. In this study, we tested a new deep learning model that uses T1-weighted (T1w) MRI scans—common clinical images—to estimate brain pathways. We compared its performance with traditional dMRI-based tractography and atlas-based methods in healthy young people, older adults, and patients with epilepsy, depression, and brain cancer. In healthy young people, our deep learning model showed slightly more error than traditional tractography, but the difference was small and less than errors seen with atlas-based methods. We also found that the model could replicate some important findings from previous dMRI studies in the clinical groups, especially for long-range brain connections that atlas methods miss, but not in all cases. These results suggest that deep learning using T1w MRI shows promise for clinical tractography, especially compared to atlas-based methods, but still needs improvement and careful testing before it can be widely used in hospitals. Additionally, our findings raise new questions about how differences between dMRI and T1w MRI scans affect tractography results, and more research on this will help us better understand what brain features influence these measurements.
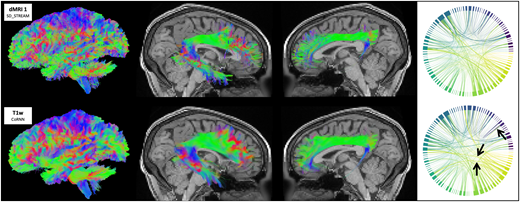
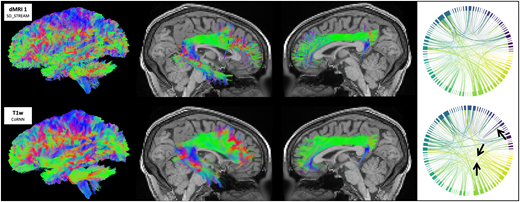
Fig 1
Tractograms (left view), right arcuate fasciculi (right view), left cinguli (left view), and cortical connectomes from traditional SD_STREAM tractography and the CoRNN method in a representative in-distribution HCP participant. Arrows denote visually appreciable differences between connectomes.